Researchers create framework to identify antibiotic-resistance genes and antibiotic-resistant bacteria in nanopore reads
Wastewater treatment plant (WWTP) effluent discharge could induce resistome enrichment in the receiving water environments. However, because of the general lack of a robust antibiotic-resistant bacteria (ARB) identification method, the driving mechanism for resistome accumulation in the receiving environment is unclear.
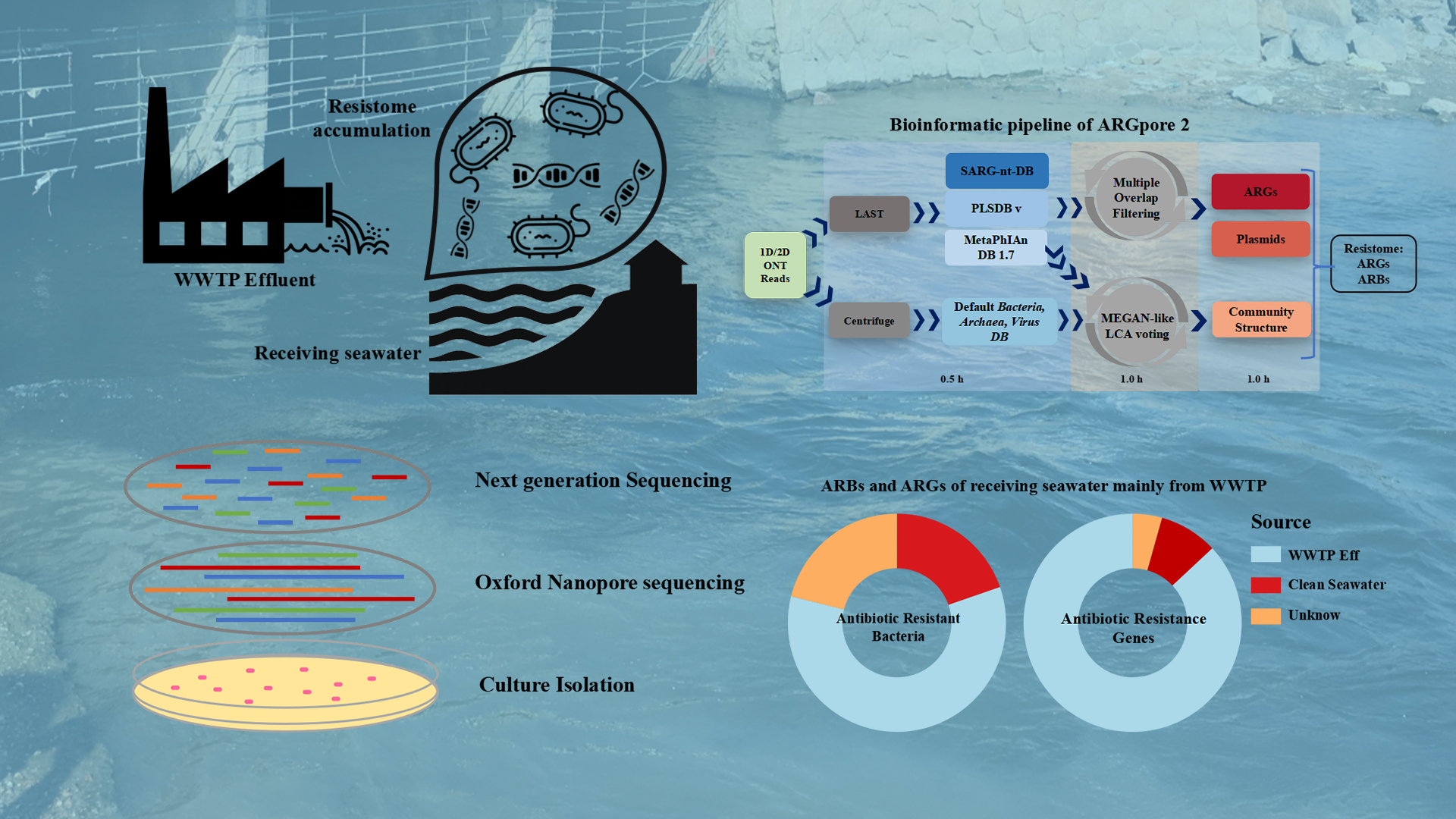
Associate Professor Yu Xia’s team from the School of Environmental Science and Engineering at the Southern University of Science and Technology (SUSTech) has recently published their research results where they distinguish the indigenous ARBs and the accumulation of WWTP-borne ARBs in the receiving water body of a domestic WWTP. A bioinformatic framework, ARGpore2, was constructed and evaluated to facilitate antibiotic resistance genes (ARGs) and ARBs identification in nanopore reads.
Their paper, entitled “Nanopore-based long-read metagenomics uncover the resistome intrusion by antibiotic resistant bacteria from treated wastewater in receiving water body,” has been published in Water Research, a high-impact journal covering all aspects of the science and technology of the anthropogenic water cycle, water quality, and its management worldwide.
To identify ARGs and ARBs in nanopore datasets, the bioinformatics framework – ARGPore2 was constructed and evaluated (Fig. 1a). As shown in Fig. 1b, ARGs identification with a similarity cut-off of 75% showed precision stably around 95%, which indicates effective filtering of false-positive hits at this cut-off. The evaluation of ARGPore2 demonstrated that nanopore-based DNA alignment is capable of predicting ARGs at an accuracy comparable to that of this widely adopted BLASTP method.
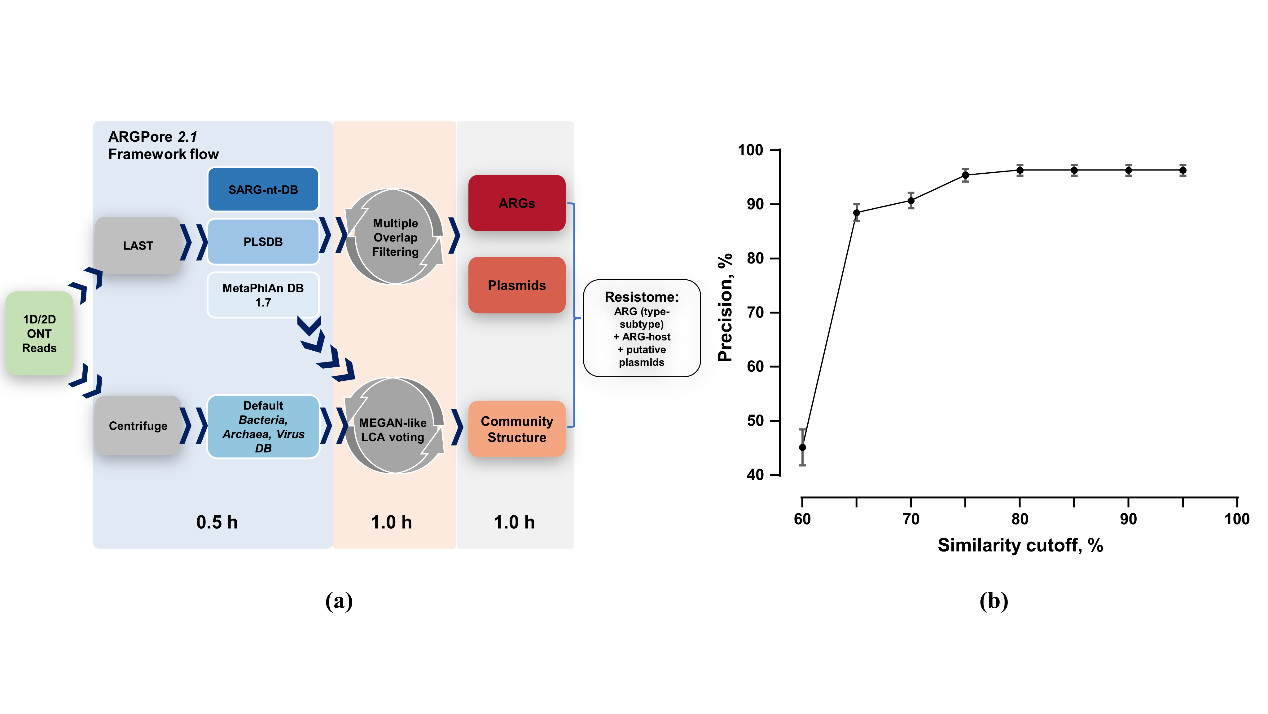
Figure 1. Workflow and performance of ARGPore2.0 (a) The analysis workflow of ARGPore2 pipeline; (b) Precision of ARGs identification at different similarity cut-offs by ARGpore2.
In the team’s Illumina-based datasets, only 18 ARGs-containing contigs are identified in 10 bacterial genera affiliated to 3 classes by metagenomic assembly accounting for 0.04% of the receiving water bacterial community, which constituted a very small fraction of resistome detected by unassembled short reads (Fig. 2a). In contrast, nanopore datasets detect 380 ARBs with phylogenetic spectrum enlarged to 33 genera affiliated to 12 classes, which was enlarged at least twofold. Additionally, culture-based validation demonstrated a high degree of congruence with the ARBs profile, which was revealed by nanopore sequencing and showed that Escherichia and Klebsiella were predominant resistant populations. At the same time, the Escherichia carrying diverse ARGs against beta-lactam were overlooked by Illumina-assembly (Fig. 2a & 2b).
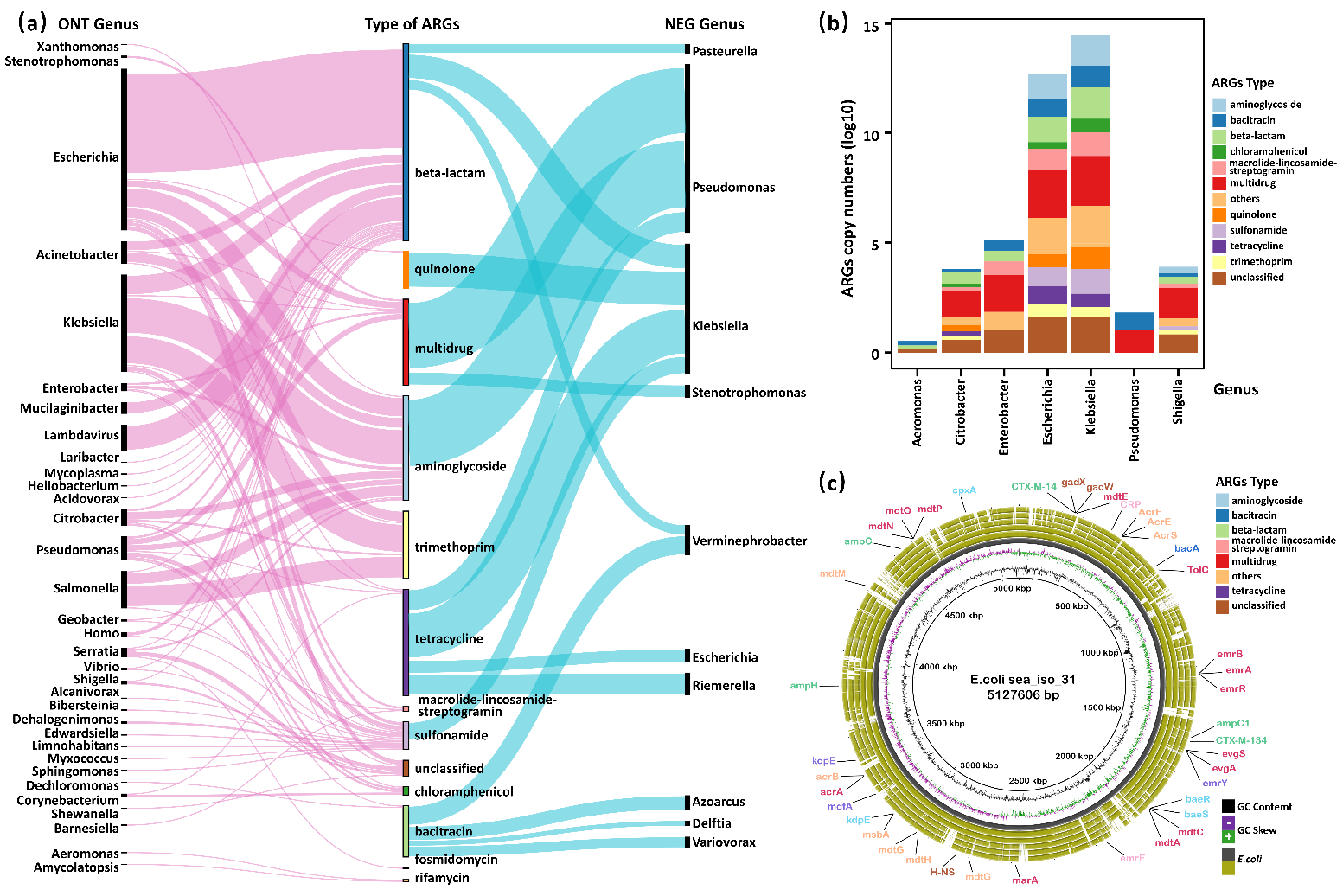
Figure 2. Comparison of ARGs host identification between nanopore-based and Illumina-assembled metagenomics. ARGs are coloured according to their resistance types. (a) The Sankey plot represents ARGs host populations identified based on nanopore and Illumina-assembly at the genus level. The width of the ribbon is proportional to the relative abundance of host populations in the corresponding dataset. (b) The ARGs profile of culture-based validation based on 24 beta-lactam-resistant bacterial isolates. (c) BRIG genome map depicting resistome of 6 isolated strains of E. coli.
A total of 65 subtypes belonging to 14 types of ARGs were detected and shown in Fig. 3a. Microbial resistance against 12 classes of antibiotics were detected in receiving seawater samples, while the researchers only identified six types in the clean seawater samples. The overall resistome concentration of receiving water community was roughly ten times higher than of clean seawater, with subtypes of TEM-group1, OXA-group2, aac(6’)-I, aadA, and dfrA7/A17 being the most sharply increased. Beta-lactam resistance genes were the most dominant resistance type in receiving water samples, primarily composed of the TEM-group and OXA-group. Furthermore, TEM-group beta-lactamases, aadA aminoglycoside, and dfrA7/A17 trimethoprim which had shown clinical relevance, were among the most prevalent resistance genotypes in receiving water samples.
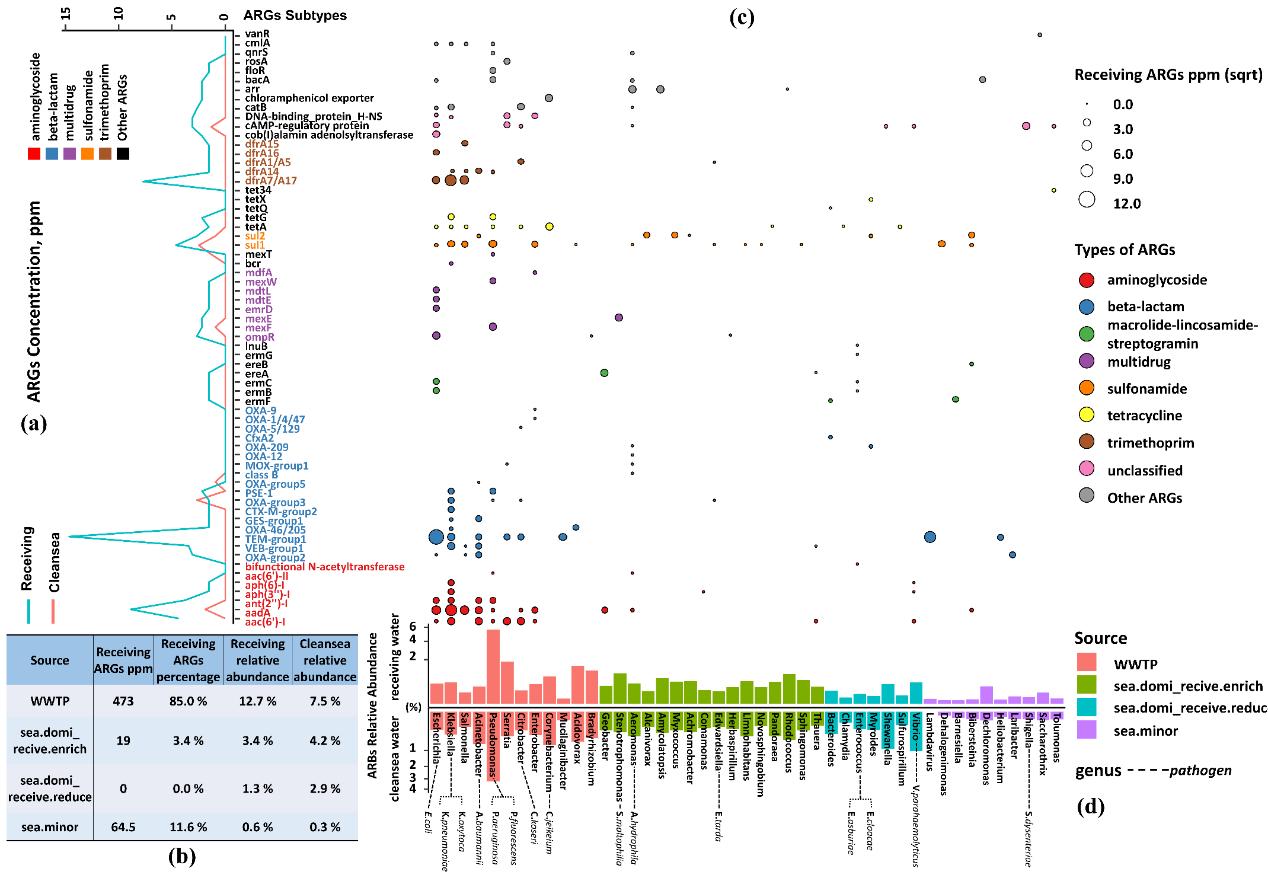
Figure 3. Profile of resistome of receiving seawater and clean seawater. (a) The comparison of the mean concentration of ARGs in receiving seawater and clean seawater samples; (b) The host range of ARGs and the fate of antibiotic resistance pathogens in coastal water systems; (c) Heatmap indicated the concentration of ARGs in receiving seawater samples; (d) Table showed that relative abundance and total concentration of ARGs of four source groups.
The potential mobility of identified ARGs were investigated by identifying the MGEs-associated genes localized on the same genetic fragment (long nanopore read). Results showed that ARGs identified in all clean seawater and receiving seawater were mostly located on bacterial chromosomes (74%) rather than on plasmids, which was contradictory to that observed in WWTP-associated resistome, including influent, activated sludge, and effluent. As for the general trend, the ARGs encoded on plasmids and chromosomes were more prevalent in receiving water samples than in clean seawater samples. Nevertheless, more than half the proportion (54%) of ARGs were encoded on chromosomes rather than plasmids in the receiving water samples, which showed a similar trend of the whole resistome distribution in all samples.

Figure 4. The distribution of ARGs carried by plasmids and chromosomes in receiving seawater samples and clean seawater samples. The left subfigure shows the overall distribution, while the differences between receiving water and clean seawater are shown in the right subfigure.
In addition to plasmids, a variety of MGEs were identified on the chromosomes. Indeed, 16 subtypes belonging to six ARG types showed close genetic association with integrons, transposons, and recombinase, implying their high tendency of HGT (Fig. 5). The genetic organization revealed by nanopore long reads showed strong connections between aac-families, aad-families and sul1 (Fig. 5), which are important components of gene cassettes of class 1 integron (CL1). Since CL1 had been reported as a clinically relevant genetic element with the ability to transfer ARGs conferring resistance to multiple antibiotics, these findings demonstrate that CL1 has played an important role in enabling HGT of chromosomal ARGs among bacteria of different phylogenetic lineages.
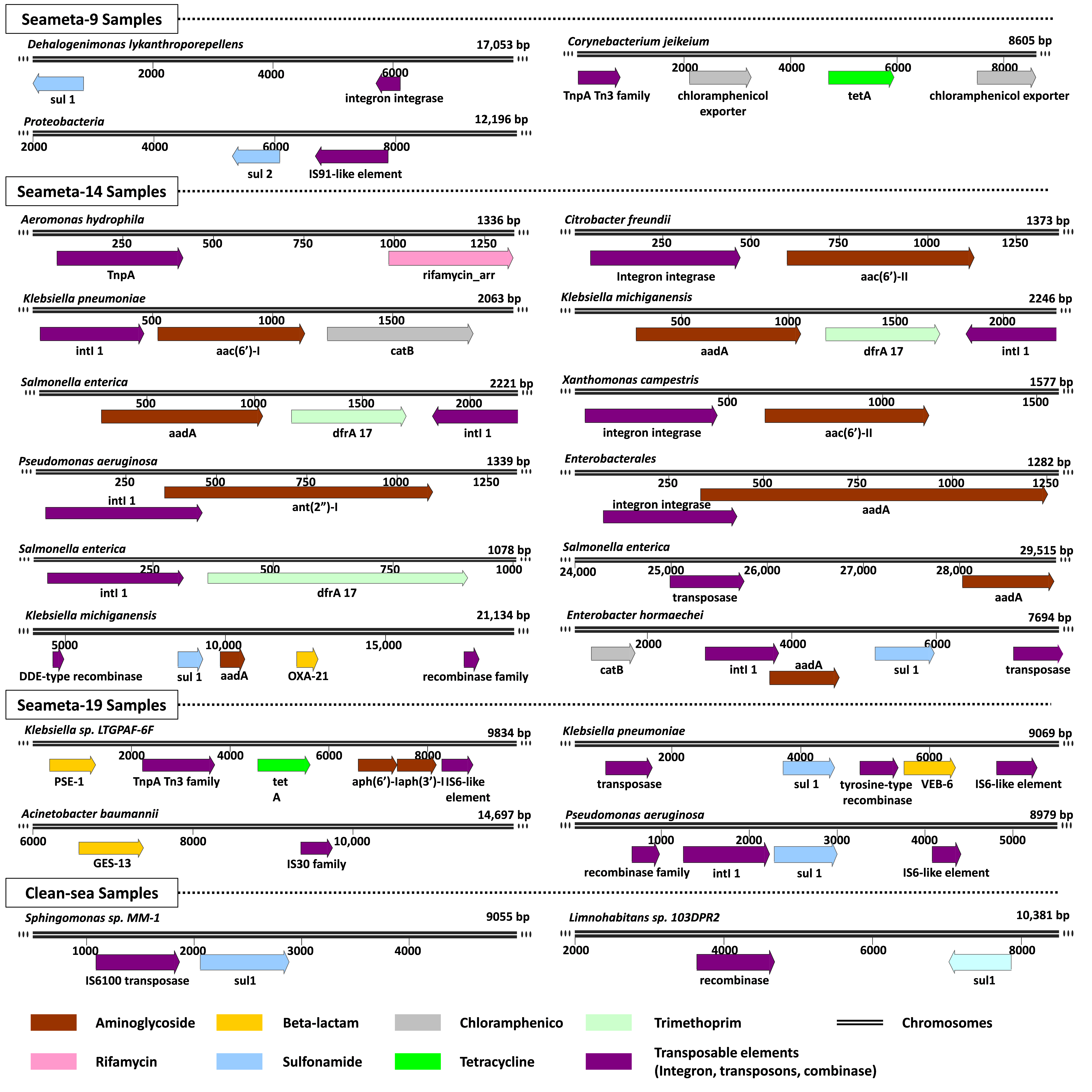
Figure 5. Schematic of the genetic organization of antibiotic resistance gene clusters identified on nanopore reads.
To understand the mechanism arousing the environmental resistome reservoir of coastal-receiving water communities, this study audaciously applied nanopore sequencing technologies to comprehensively investigate the dissemination and enrichment of ARBs. A robust bioinformatic framework (ARGpore2) was constructed and carefully tested to predict resistance and establish plasmids/MGE-associated mobility.
Prof. Xia’s team results suggest that the receiving coastal system might be considered as a reservoir for ARGs mediated by the robust installation of WWTP-borne ARB community coupled with recombination and HGT enabled by plasmids and class 1 integrons. Consequently, in order to mitigate resistance pollution in receiving environments, advanced filtration or oxidative process should be included in the disinfection step of WWTPs to enable more complete disruption of pathogen cells and endospores during treatment.
Ziqi Wu, a joint Ph.D. student of SUSTech and the University of Copenhagen, is the first author of this paper. Assoc. Prof. Yu Xia at SUSTech is the corresponding author. Prof. Tong Zhang’s team from the University of Hong Kong (HKU) also contributed to this study.
This work was supported by the National Natural Science Foundation of China (NSFC).
Paper link: https://doi.org/10.1016/j.watres.2022.119282
ARGPore2 link: https://github.com/sustc-xylab/ARGpore2
